Subtopic 4.6: Expression Array Assay
The expression array is the chip based microarray of more gene expressions (finger print of genes) by the effect of cellular toxicants. This is a rapid and sensitive detection method which allows to detect all toxicological end points at wide range of molecular level changes in the cell at single assay. The microarray process can be divided into two main parts. First is the printing of known gene sequences onto glass slides or other solid support followed by hybridization of fluorescently labeled cDNA (containing the unknown sequences to be interrogated) to the known genes immobilized on the glass slide. After hybridization, arrays are scanned using a fluorescent microarray scanner. Analyzing the relative fluorescent intensity of different genes provides a measure of the differences in gene expression (Figure 6).
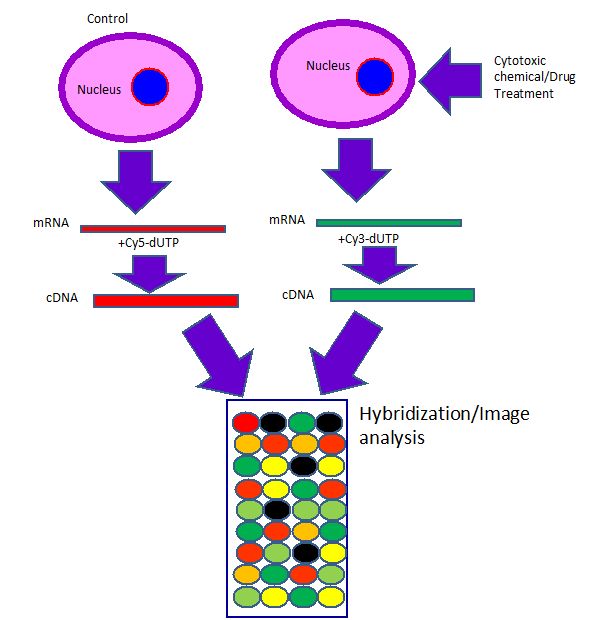
Figure 6. Schematic representation of gene expression array showed that Cy5(red) and Cy3 (green) labeled cDNA hybridized to a DNA microarray. Yellow spots indicate the genes are expressed in both samples. The intensity and different types of color at each spot indicate the level and presence of genes in samples. Black spots show low level of expression or do not show any expression of genes.



