Subtopic 3.2: Genetic Mutation Assay
Following are the different molecular assay to study nucleotide variants or alternation of genetic material caused by mutagens:
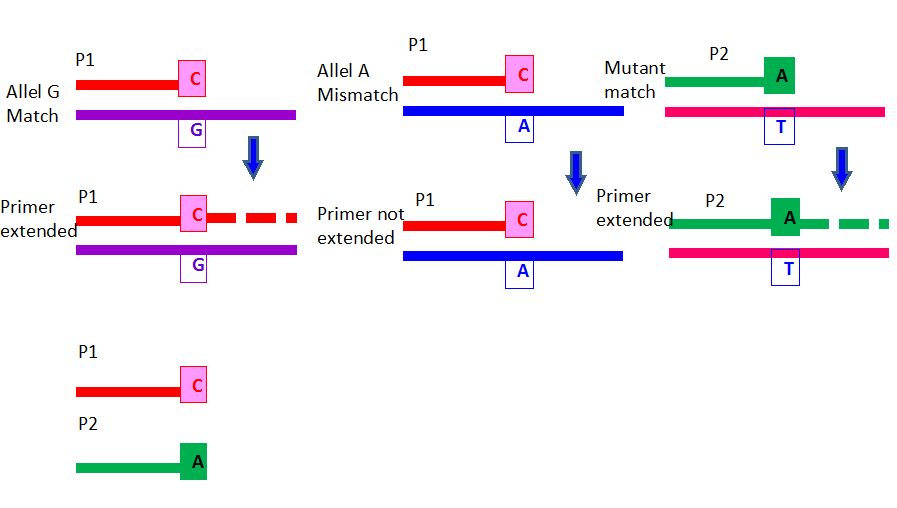
3.2a. Allele-Specific PCR :
- Single nucleotide polymorphism (SNP) resulted from base substitution mutation can be analyzed by this method. In this real-time PCR, fluorescent reporter probes are added to the reaction mixture and one fluorescent reporter probe is selected for wild type and other fluorescent probe is used for mutant. The PCR primers with fluorescent probe will match or mismatch one of the alleles at 3’ end of the primer. DNA polymerase extends the probes in a complementary fashion and releasing the reporter fluorescent molecules for detection. The PCR cycles with the reporter probes show the amplified signals and allow for precise measurement of one or both alleles of interest. Similarly, the 3’ end of the mutant-specific primer is extended only in the presence of DNA with that mutation (Figure 2).
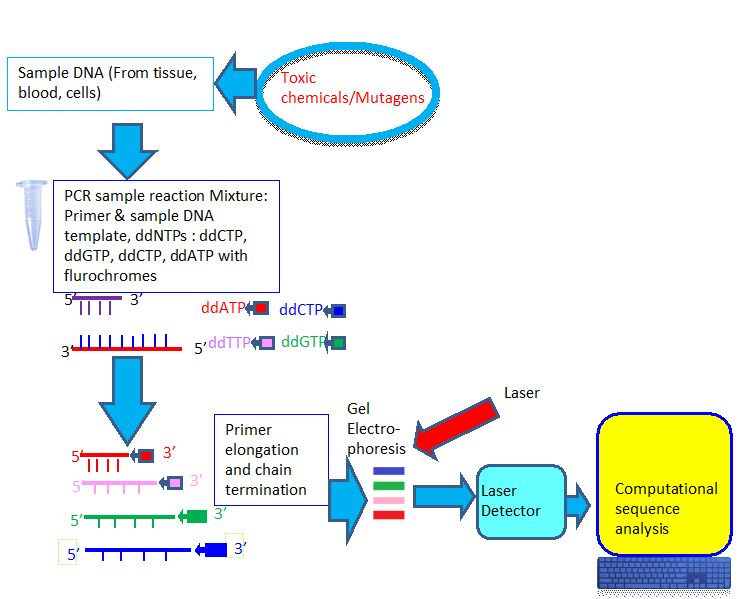
3.2b. Sanger Dideoxy Sequencing :
- The goal of this method is to detect unknown mutations including single nucleotide variants (SNVs) and small duplications, insertions, deletions, and indels of interest caused by mutagens. In this method, sequencing primers hybridized to the PCR product and are extended using the four deoxynucleotides (dNTPs), a mixture of fluorescently labeled dideoxynucleotides (ddNTPs) and DNA polymerase. Four ddNTPs are marked with a different fluorescent dye. Random incorporation of the marked ddNTPs shows in termination of strands at each location along the sequence. The gel electrophoresis separates the strands by size. Fluorescence spectroscopy measured the terminating nucleotides (Figure 3).